Custom model and Transform¶
This notebook contains the simple examples of custom model and Transform that can be added to the ETNA framework.
Table of Contents
[1]:
import warnings
from pandas.errors import SettingWithCopyWarning
warnings.filterwarnings(action="ignore", message="Torchmetrics v0.9")
warnings.filterwarnings(action="ignore", message="`tsfresh` is not available")
warnings.filterwarnings(action="ignore", message="Using categorical_feature")
warnings.filterwarnings(action="ignore", message="Argument `closed` is deprecated")
warnings.filterwarnings(action="ignore", message="Using the level keyword")
warnings.filterwarnings(action="ignore", category=SettingWithCopyWarning)
[2]:
import numpy as np
import pandas as pd
from etna.datasets.tsdataset import TSDataset
from etna.transforms import LagTransform
from etna.transforms import SegmentEncoderTransform
from etna.transforms import DateFlagsTransform
from etna.transforms import LinearTrendTransform
from etna.pipeline import Pipeline
from etna.metrics import MAE
from etna.analysis import plot_backtest
1. What is Transform and how it works¶
Our library works with the special data structure TSDataset. So, before starting, we need to convert the classical DataFrame to TSDataset.
[3]:
df = pd.read_csv("data/example_dataset.csv")
df["timestamp"] = pd.to_datetime(df["timestamp"])
df = TSDataset.to_dataset(df)
ts = TSDataset(df, freq="D")
ts.head(5)
[3]:
| segment | segment_a | segment_b | segment_c | segment_d |
|---|---|---|---|---|
| feature | target | target | target | target |
| timestamp | ||||
| 2019-01-01 | 170 | 102 | 92 | 238 |
| 2019-01-02 | 243 | 123 | 107 | 358 |
| 2019-01-03 | 267 | 130 | 103 | 366 |
| 2019-01-04 | 287 | 138 | 103 | 385 |
| 2019-01-05 | 279 | 137 | 104 | 384 |
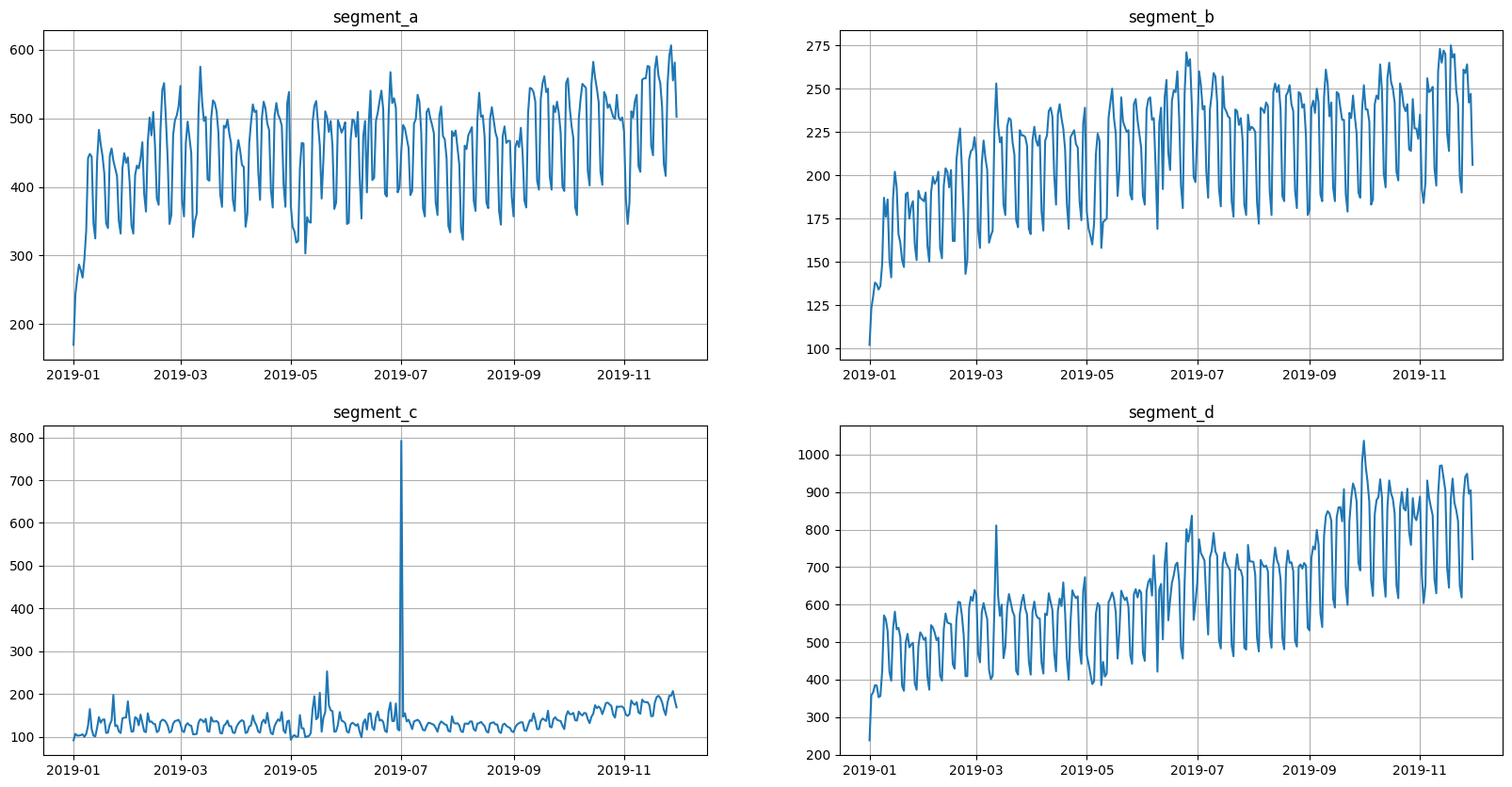
Let’s look at the original view of data
[4]:
ts.plot()
Transform is the manipulation of data to extract new features or update created ones.
In ETNA, Transforms can change column values or add new ones.
For example:
DateFlagsTransform - adds columns with information about the date (day number, is the day a weekend, etc.) .
LinearTrendTransform - subtracts a linear trend from the series (changes it).
[5]:
dates = DateFlagsTransform(day_number_in_week=True, day_number_in_month=False, out_column="dateflag")
detrend = LinearTrendTransform(in_column="target")
ts.fit_transform([dates, detrend])
ts.head(3)
[5]:
| segment | segment_a | segment_b | segment_c | segment_d | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| feature | dateflag_day_number_in_week | dateflag_is_weekend | target | dateflag_day_number_in_week | dateflag_is_weekend | target | dateflag_day_number_in_week | dateflag_is_weekend | target | dateflag_day_number_in_week | dateflag_is_weekend | target |
| timestamp | ||||||||||||
| 2019-01-01 | 1 | False | -236.276825 | 1 | False | -79.162964 | 1 | False | -26.743498 | 1 | False | -194.070140 |
| 2019-01-02 | 2 | False | -163.575877 | 2 | False | -58.358457 | 2 | False | -11.861383 | 2 | False | -75.292679 |
| 2019-01-03 | 3 | False | -139.874928 | 3 | False | -51.553950 | 3 | False | -15.979267 | 3 | False | -68.515217 |
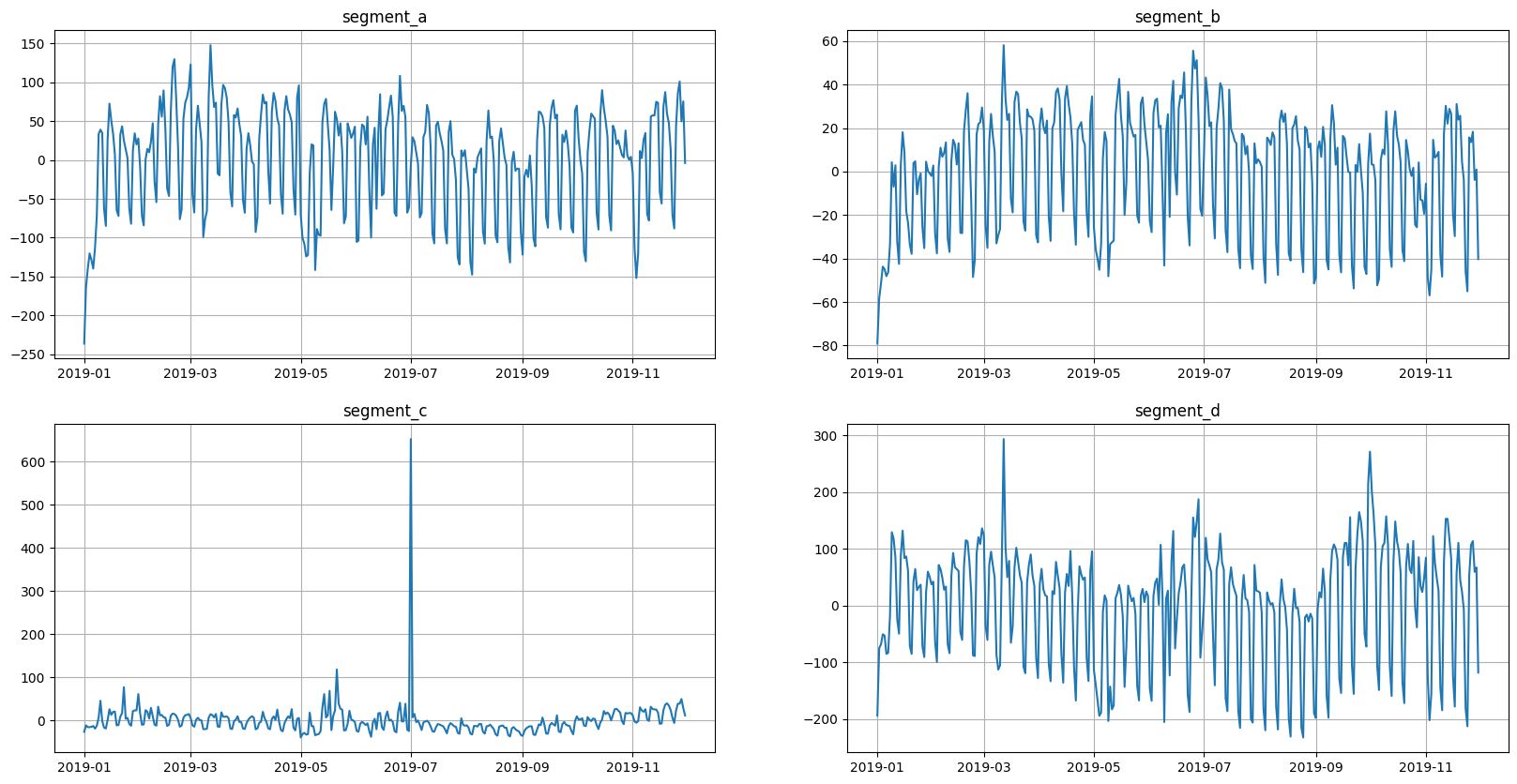
In addition to the appearance of a new column, the values in the target column have changed. This can be seen from the graphs.
[6]:
ts.plot()
[7]:
ts.inverse_transform([dates, detrend])
ts.head(3)
[7]:
| segment | segment_a | segment_b | segment_c | segment_d | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| feature | dateflag_day_number_in_week | dateflag_is_weekend | target | dateflag_day_number_in_week | dateflag_is_weekend | target | dateflag_day_number_in_week | dateflag_is_weekend | target | dateflag_day_number_in_week | dateflag_is_weekend | target |
| timestamp | ||||||||||||
| 2019-01-01 | 1 | False | 170.0 | 1 | False | 102.0 | 1 | False | 92.0 | 1 | False | 238.0 |
| 2019-01-02 | 2 | False | 243.0 | 2 | False | 123.0 | 2 | False | 107.0 | 2 | False | 358.0 |
| 2019-01-03 | 3 | False | 267.0 | 3 | False | 130.0 | 3 | False | 103.0 | 3 | False | 366.0 |
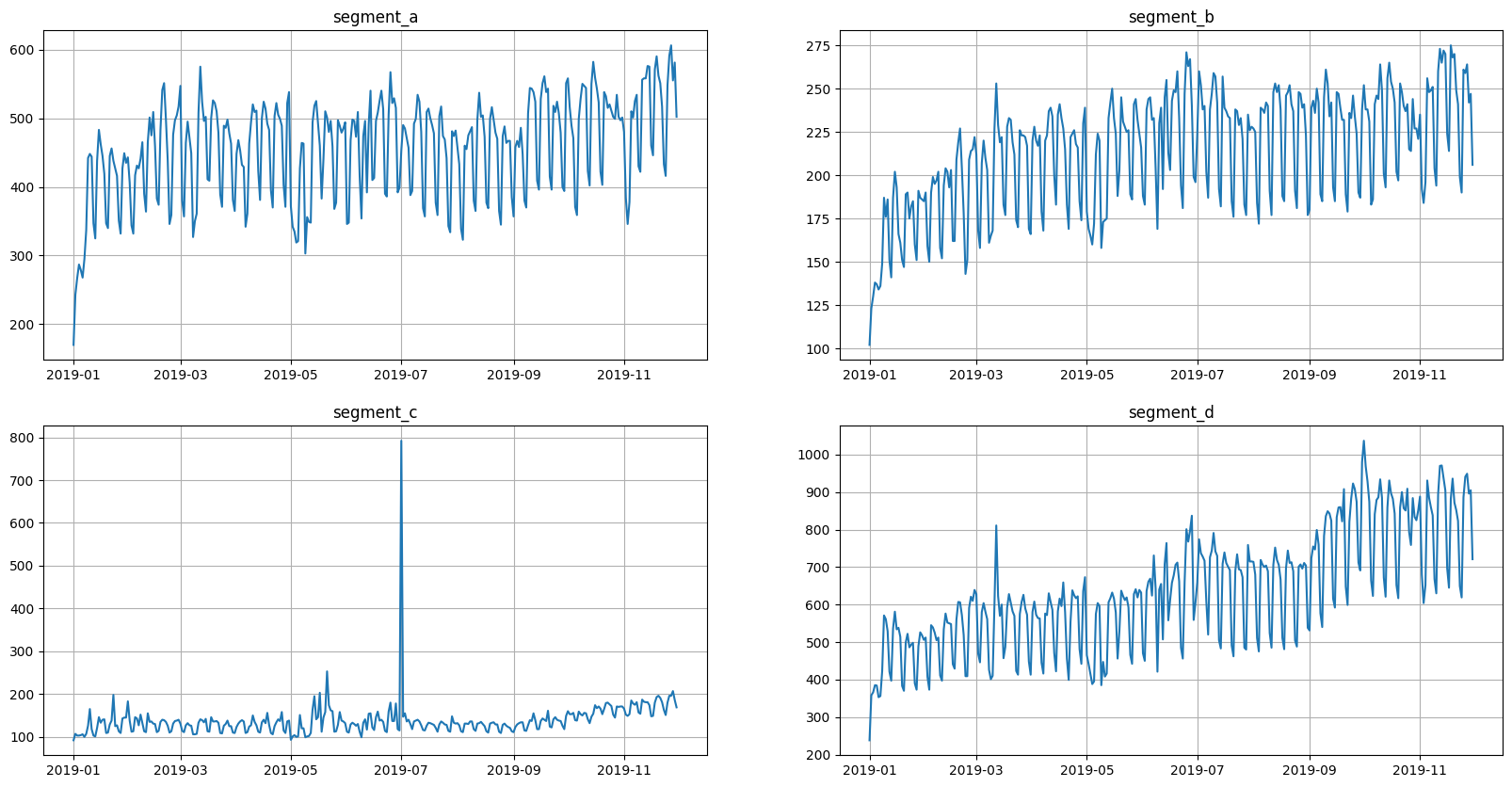
Now the data is back in its original form
[8]:
ts.plot()
2. Custom Transform¶
Let’s define custom Transform.
Consider a Transform that sets bounds at the top and bottom - FloorCeilTransform
2.1 Per-segment Custom Transform¶
Consider the implementation of this Transform in per-segment way.
First of all, we need to implement logic of the Transform for one segment. To do so, we need to inherit from OneSegmentTransform and implement all its methods.
[9]:
from etna.transforms.base import OneSegmentTransform
[10]:
# Class for processing one segment.
class _OneSegmentFloorCeilTransform(OneSegmentTransform):
# Constructor with the name of the column to which the transformation will be applied.
def __init__(self, in_column: str, floor: float, ceil: float):
"""
Create instance of _OneSegmentLinearTrendBaseTransform.
Parameters
----------
in_column:
name of processed column
floor:
lower bound
ceil:
upper bound
"""
self.in_column = in_column
self.floor = floor
self.ceil = ceil
# Provide the necessary training. For example calculates the coefficients of a linear trend.
# In this case, we calculate the indices that need to be changed
# and remember the old values for inverse transform.
def fit(self, df: pd.DataFrame) -> "_OneSegmentFloorCeilTransform":
"""
Calculate the indices that need to be changed.
Returns
-------
self
"""
target_column = df[self.in_column]
self.floor_indices = target_column < self.floor
self.floor_values = target_column[self.floor_indices]
self.ceil_indices = target_column > self.ceil
self.ceil_values = target_column[self.ceil_indices]
return self
# Apply changes.
def transform(self, df: pd.DataFrame) -> pd.DataFrame:
"""
Drive the value to the interval [floor, ceil].
Parameters
----------
df:
DataFrame to transform
Returns
-------
transformed series
"""
result_df = df
result_df[self.in_column].iloc[self.floor_indices] = self.floor
result_df[self.in_column].iloc[self.ceil_indices] = self.ceil
return result_df
# Returns back changed values.
def inverse_transform(self, df: pd.DataFrame) -> pd.DataFrame:
"""
Inverse transformation for transform. Return back changed values.
Parameters
----------
df:
data to transform
Returns
-------
pd.DataFrame
reconstructed data
"""
result = df
result[self.in_column][self.floor_indices] = self.floor_values
result[self.in_column][self.ceil_indices] = self.ceil_values
return result
Now we can define a class, which will work with the entire dataset, applying a transform(_OneSegmentFloorCeilTransform) to each segment.
This functionality is provided by PerSegmentWrapper. There are two versions of it: 1. IrreversiblePerSegmentWrapper — base class for per-segment transforms without inverse transformation. This class implements the inverse_transform simply returning the entire dataset. 2. ReversiblePerSegmentWrapper — base class for per-segment transforms with custom inverse transformation. This class implements the inverse_transform logic calling the corresponding method of
OneSegmentTransform inside for each segment.
The one thing we need to implement in both cases is only the method get_regressors_info — it should return the regressors, created by the Transform.
[11]:
from etna.transforms.base import ReversiblePerSegmentWrapper
from typing import List
[12]:
class FloorCeilPerSegmentTransform(ReversiblePerSegmentWrapper):
"""Transform that truncate values to an interval [ceil, floor]"""
def __init__(self, in_column: str, floor: float, ceil: float):
"""Create instance of FloorCeilTransform.
Parameters
----------
in_column:
name of processed column
floor:
lower bound
ceil:
upper bound
"""
self.in_column = in_column
self.floor = floor
self.ceil = ceil
super().__init__(
transform=_OneSegmentFloorCeilTransform(in_column=self.in_column, floor=self.floor, ceil=self.ceil),
required_features=[in_column],
)
# Here we need to specify output columns with regressors, if transform creates them.
def get_regressors_info(self) -> List[str]:
"""Return the list with regressors created by the transform.
Returns
-------
:
List with regressors created by the transform.
"""
return []
2.2 Multi-segment Custom Transform¶
Now, consider the implementation of this Transform in multi-segment way.
For multi-segment Transforms we have the similar separation for two base classes: 1. IrreversibleTransform — base class for multi-segment transforms without inverse transformation. This class implements the inverse_transform simply returning the entire dataset. The other logic should be implemented in _fit and _transform methods. 2. ReversibleTransform — base class for multi-segment transforms with custom inverse transformation. In addition to _fit, _transform here you
should implement the logic of inverse transformation in method _inverse_transform.
Methods you implement work with the dataframe, the logic of processing the TSDataset is implemented in the public versions of these methods, which work with TSDataset now.
[13]:
from etna.transforms.base import ReversibleTransform
[14]:
# Class for processing one segment.
class FloorCeilMultiSegmentTransform(ReversibleTransform):
# Constructor with the name of the column to which the transformation will be applied.
def __init__(self, in_column: str, floor: float, ceil: float):
"""
Create instance of FloorCeilMultiSegmentTransform.
Parameters
----------
in_column:
name of processed column
floor:
lower bound
ceil:
upper bound
"""
super().__init__(required_features=[in_column]) # only these features will be passed to the other methods
self.in_column = in_column
self.floor = floor
self.ceil = ceil
# Provide the necessary training. For example calculates the coefficients of a linear trend.
# In this case, we calculate the indices that need to be changed
# and remember the old values for inverse transform.
def _fit(self, df: pd.DataFrame) -> "FloorCeilMultiSegmentTransform":
"""
Calculate the indices that need to be changed.
Returns
-------
self
"""
target_column = df.loc[pd.IndexSlice[:], pd.IndexSlice[:, self.in_column]]
self.floor_indices = target_column < self.floor
self.floor_values = target_column[self.floor_indices]
self.ceil_indices = target_column > self.ceil
self.ceil_values = target_column[self.ceil_indices]
return self
# Apply changes.
def _transform(self, df: pd.DataFrame) -> pd.DataFrame:
"""
Drive the value to the interval [floor, ceil].
Parameters
----------
df:
DataFrame to transform
Returns
-------
transformed series
"""
result_df = df
result_df[self.floor_indices] = self.floor
result_df[self.ceil_indices] = self.ceil
return result_df
# Returns back changed values.
def _inverse_transform(self, df: pd.DataFrame) -> pd.DataFrame:
"""
Inverse transformation for transform. Return back changed values.
Parameters
----------
df:
data to transform
Returns
-------
pd.DataFrame
reconstructed data
"""
result_df = df
result_df[self.floor_indices] = self.floor_values[self.floor_indices]
result_df[self.ceil_indices] = self.ceil_values[self.ceil_indices]
return result_df
# Here we need to specify output columns with regressors, if transform creates them.
def get_regressors_info(self) -> List[str]:
"""Return the list with regressors created by the transform.
Returns
-------
:
List with regressors created by the transform.
"""
return []
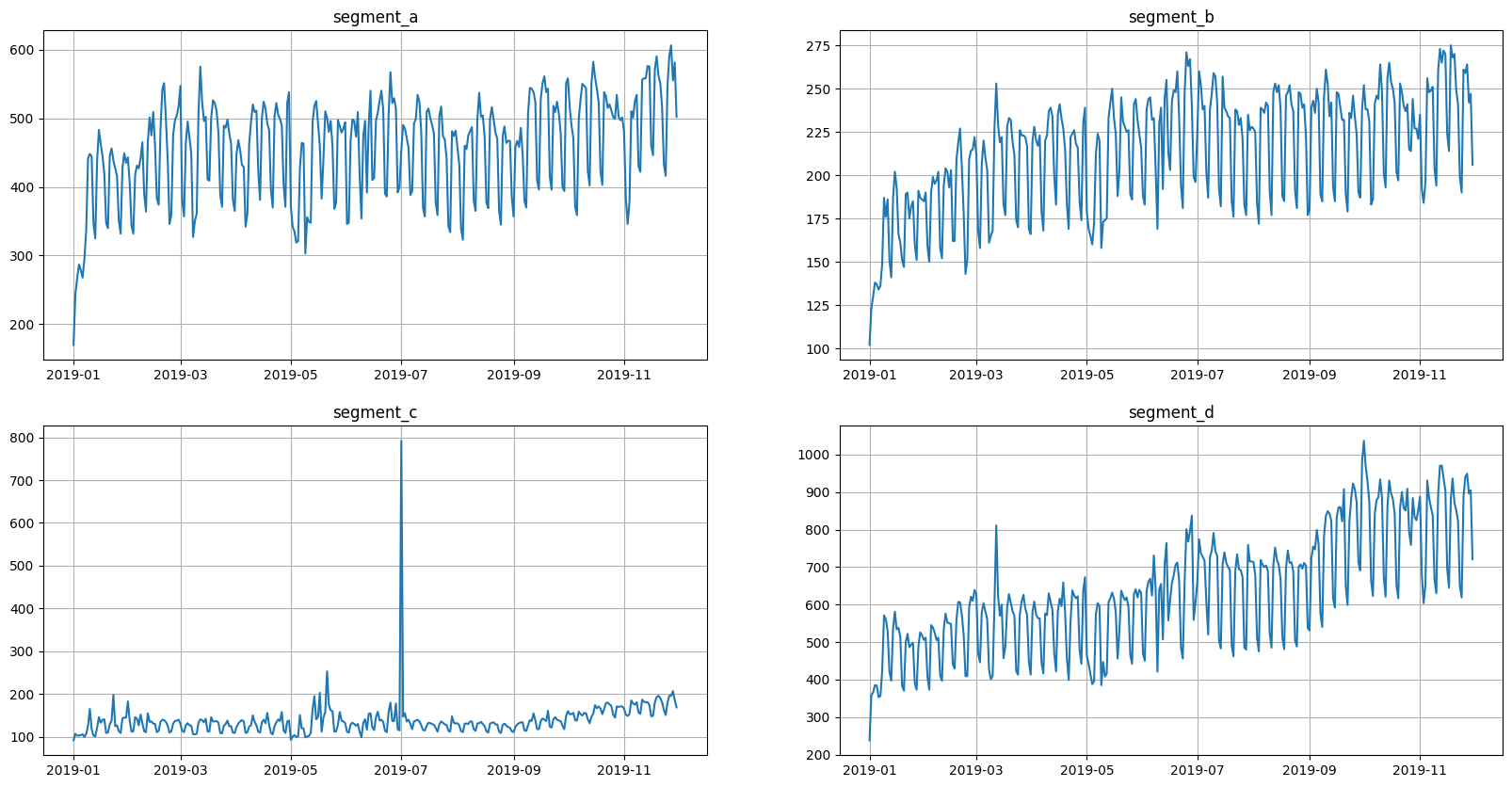
Lets take a closer look.
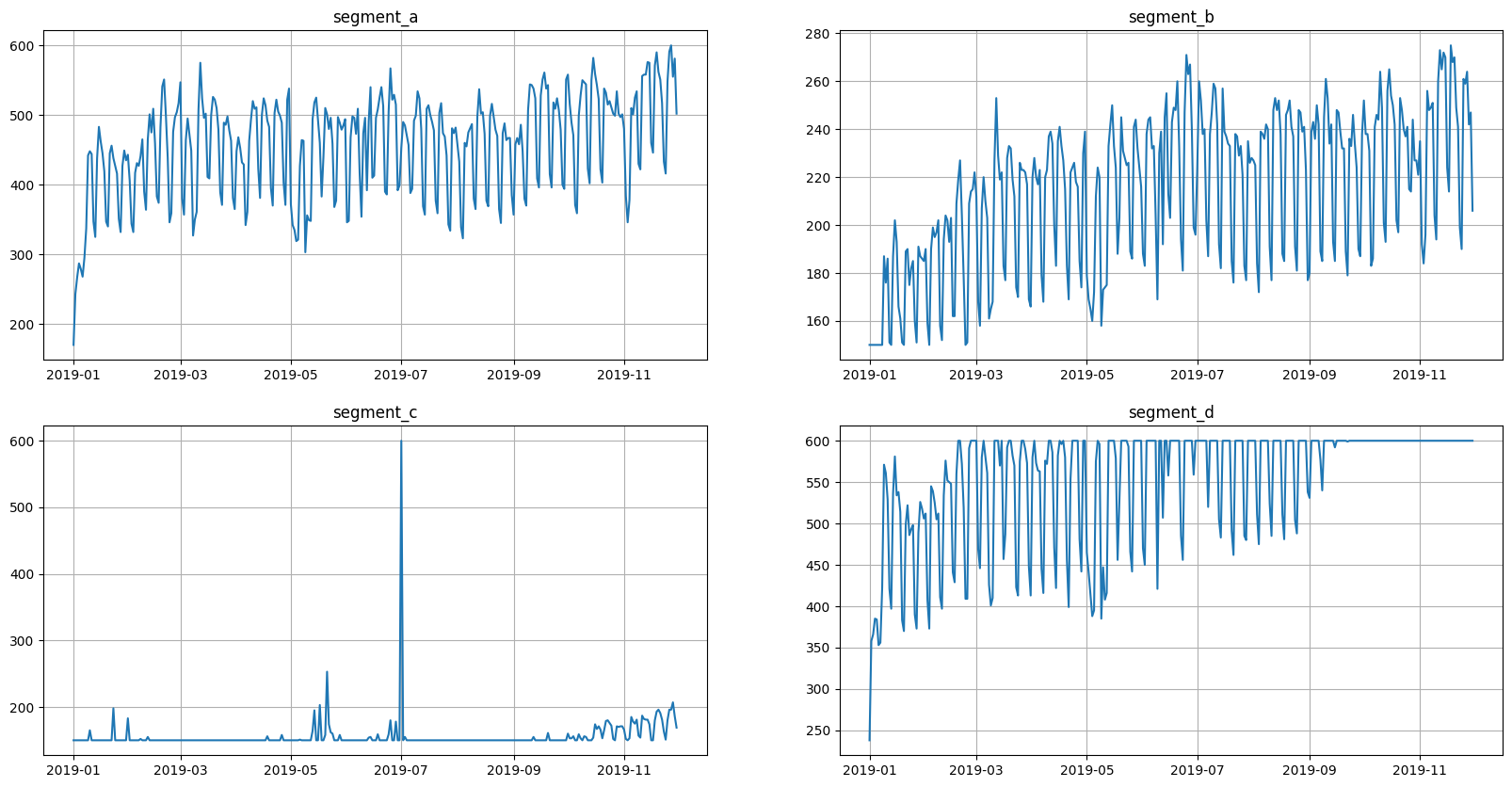
This is what the original data looks like.
[15]:
ts.plot()
Let’s check that our implementations are equivalent
[16]:
from copy import deepcopy
[17]:
bounds_multi_segment = FloorCeilMultiSegmentTransform(in_column="target", floor=150, ceil=600)
bounds_per_segment = FloorCeilMultiSegmentTransform(in_column="target", floor=150, ceil=600)
df_per_segment = bounds_per_segment.fit_transform(deepcopy(ts)).to_pandas()
df_multi_segment = bounds_multi_segment.fit_transform(deepcopy(ts)).to_pandas()
pd.testing.assert_frame_equal(df_per_segment, df_multi_segment)
Fine, then we can check out the work of any of them
[18]:
ts.fit_transform([bounds_multi_segment])
[19]:
ts.plot()
Returning to the original values
[20]:
ts.inverse_transform([bounds_multi_segment])
[21]:
ts.plot()
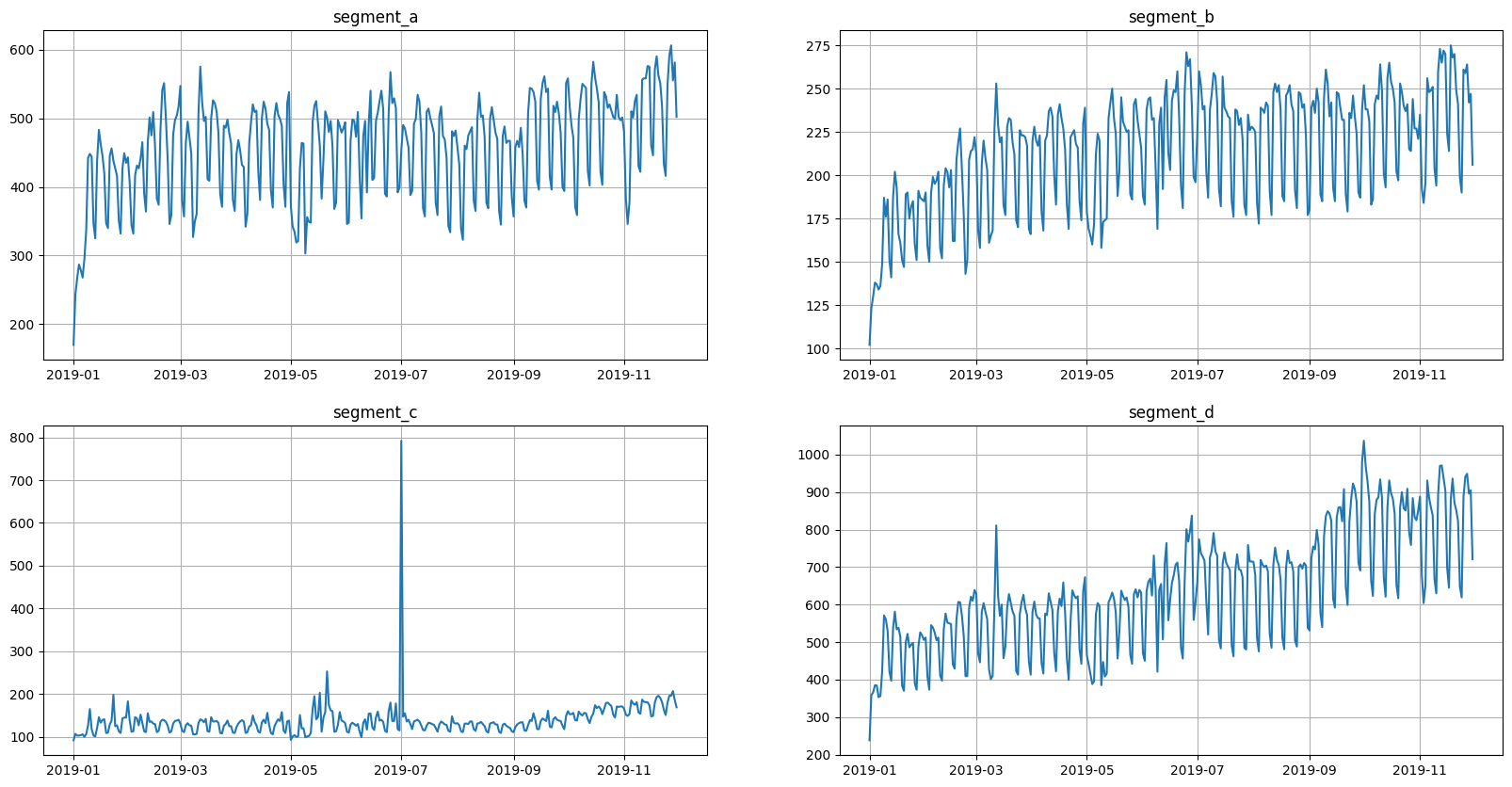
Everything seems to be working correctly. Remember to write the necessary tests before adding a new transform to the library.
3. Custom Model¶
If you could not find a suitable model among the ready-made ones, then you can create your own.
In this example we will try to add model based on lightgbm package.
[22]:
!pip install lightgbm -q
Creating a new model from scratch¶
First, let’s look at creating a new model from scratch. First of all, we should choose our base class. There are: * NonPredictionIntervalContextIgnorantAbstractModel: model can’t generate prediction intervals and doesn’t require context to make predictions, * NonPredictionIntervalContextRequiredAbstractModel: model can’t generate prediction intervals and requires context to make predictions, * PredictionIntervalContextIgnorantAbstractModel: model can generate prediction intervals
and doesn’t require context to make predictions, * PredictionIntervalContextRequiredAbstractModel: model can generate prediction intervals and requires context to make predictions.
These classes have different signatures for forecast and predict methods depending on their name. * All signatures accept ts: TSDataset parameter for making prediction and return_components: bool, that denotes whether to estimate decomposition for predictions. * If a model can generate prediction intervals it also accepts prediction_interval: bool and quantiles: Sequence[float] parameters. * If a model requires context it also accepts prediction_size: int
parameter, that is required to distinguish history context from points we want to make prediction on.
Let’s make some clarifications about the context. It is a part of a dataset before prediction points that is necessary for making forecasts. It is necessary for models that in its core use previous points to make predictions into the future. The example is etna.models.NaiveMode(lag=1) that uses last point to predict the next.
Ok, what about model based on lightgbm? This model doesn’t require context and we will make implementation that doesn’t generate prediction intervals. Also, this section shows how one can implement prediction decomposition.
[23]:
from lightgbm import LGBMRegressor
from etna.models.base import NonPredictionIntervalContextIgnorantAbstractModel
Let’s look at implementation.
[24]:
class LGBMModel(NonPredictionIntervalContextIgnorantAbstractModel):
def __init__(
self,
boosting_type="gbdt",
num_leaves=31,
max_depth=-1,
learning_rate=0.1,
n_estimators=100,
**kwargs,
):
self.boosting_type = boosting_type
self.num_leaves = num_leaves
self.max_depth = max_depth
self.learning_rate = learning_rate
self.n_estimators = n_estimators
self.kwargs = kwargs
self.model = LGBMRegressor(
boosting_type=self.boosting_type,
num_leaves=self.num_leaves,
max_depth=self.max_depth,
learning_rate=self.learning_rate,
n_estimators=self.n_estimators,
**self.kwargs,
)
def fit(self, ts: TSDataset) -> "LGBMModel":
"""Fit model.
Parameters
----------
ts:
Dataset with features
Returns
-------
:
Model after fit
"""
df = ts.to_pandas(flatten=True)
df = df.dropna()
features = df.drop(columns=["timestamp", "segment", "target"])
self._categorical = features.select_dtypes(include=["category"]).columns.to_list()
target = df["target"]
self.model.fit(X=features, y=target, categorical_feature=self._categorical)
def forecast(self, ts: TSDataset, return_components: bool = False) -> TSDataset:
"""Make predictions.
Prediction decomposition is based on SHAP values for LGBM.
Parameters
----------
ts:
Dataset with features
return_components:
If True additionally returns prediction components
Returns
-------
:
Dataset with predictions
"""
horizon = len(ts.df)
df = ts.to_pandas(flatten=True)
features = df.drop(columns=["timestamp", "segment", "target"])
y_flat = self.model.predict(features)
y = y_flat.reshape(-1, horizon).T
ts.loc[:, pd.IndexSlice[:, "target"]] = y
if return_components:
ts = self.forecast_components(ts=ts)
return ts
def forecast_components(self, ts: TSDataset) -> TSDataset:
"""Estimate prediction decomposition using SHAP values.
Parameters
----------
ts:
Dataset with features
Returns
-------
:
Dataset with predictions
"""
df = ts.to_pandas(flatten=True)
features = df.drop(columns=["timestamp", "segment", "target"])
# estimate SHAP values for prediction decomposition
shap_values = self.model.predict(features, pred_contrib=True)
# encapsulate expected contribution into components
components = shap_values[:, :-1] + shap_values[:, -1, np.newaxis] / (shap_values.shape[1] - 1)
# components names should start with prefix `target_component_`
component_names = [f"target_component_{name}" for name in features.columns]
components_df = pd.DataFrame(data=components, columns=component_names)
components_df["timestamp"] = df["timestamp"]
components_df["segment"] = df["segment"]
components_df = TSDataset.to_dataset(df=components_df)
# adding estimated components to dataset with predictions
ts.add_target_components(target_components_df=components_df)
return ts
def predict(self, ts: TSDataset, return_components: bool = False) -> TSDataset:
"""Make predictions.
Parameters
----------
ts:
Dataset with features
return_components:
If True additionally returns prediction components
Returns
-------
:
Dataset with predictions
"""
return self.forecast(ts=ts, return_components=return_components)
def get_model(self) -> LGBMRegressor:
"""Get internal lightgbm model.
Returns
-------
:
lightgbm model.
"""
return self.model
Let’s test it.
[25]:
HORIZON = 31
[26]:
trend = LinearTrendTransform(in_column="target")
lags = LagTransform(in_column="target", lags=list(range(31, 96, 1)), out_column="lag")
date_flags = DateFlagsTransform(
day_number_in_week=True,
day_number_in_month=True,
week_number_in_month=True,
week_number_in_year=True,
month_number_in_year=True,
year_number=True,
special_days_in_week=[5, 6],
out_column="dateflag",
)
segment_encoder = SegmentEncoderTransform()
transforms = [
trend,
lags,
date_flags,
segment_encoder,
]
[27]:
model = LGBMModel(random_state=42)
pipeline = Pipeline(model=model, transforms=transforms, horizon=HORIZON)
metrics_df, forecast_df, _ = pipeline.backtest(ts=ts, metrics=[MAE()], n_folds=3)
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.2s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.5s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.9s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.9s finished
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.1s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.3s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.5s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.5s finished
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.0s finished
Let’s look at the results.
[28]:
plot_backtest(forecast_df=forecast_df, ts=ts, history_len=50)
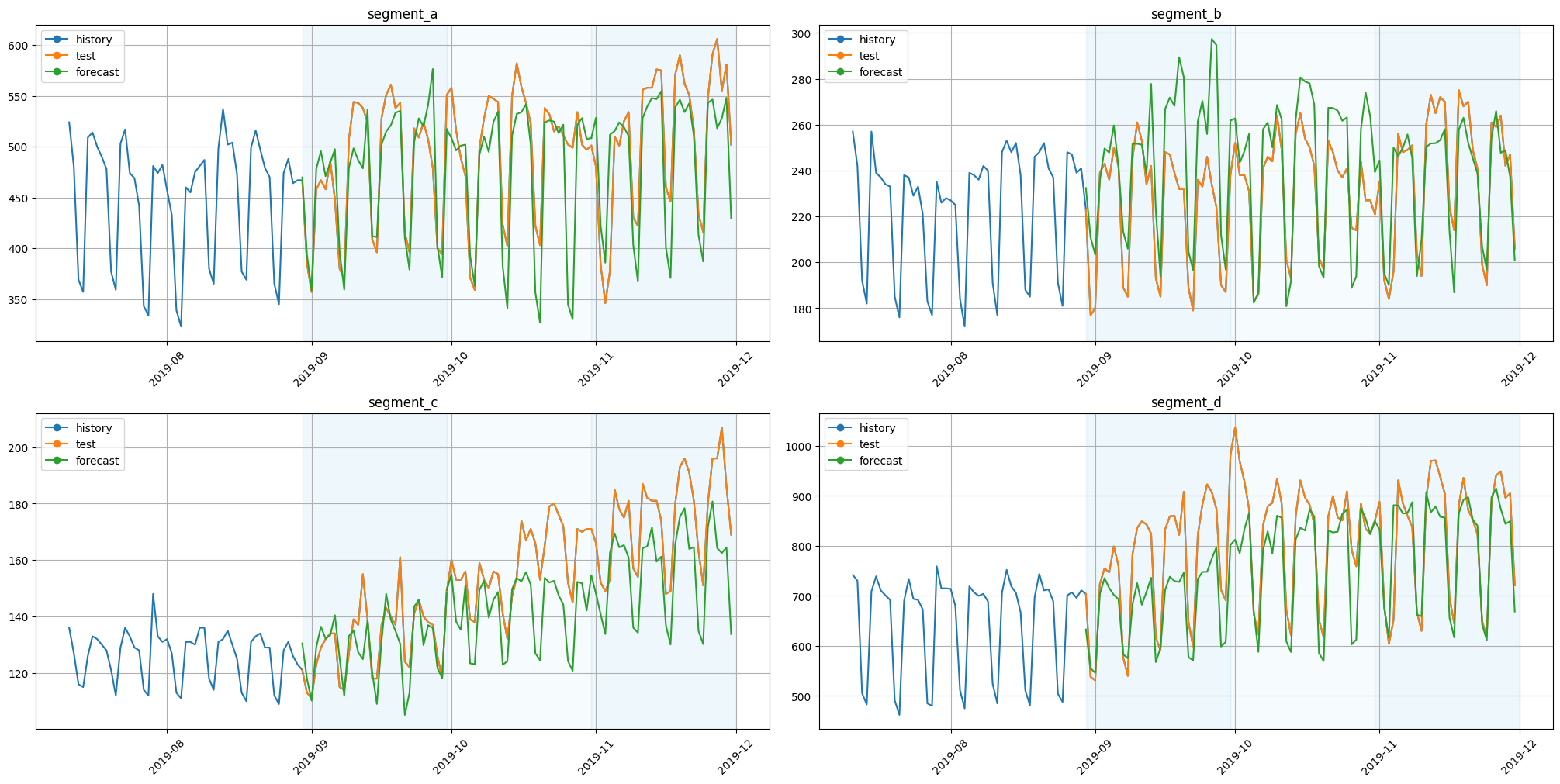
As we can see, predictions make sense.
Let’s create a pipeline with only lag features to show how prediction components could be estimated and visualised.
[29]:
from etna.analysis.forecast import plot_forecast_decomposition
model = LGBMModel(random_state=42)
pipeline = Pipeline(
transforms=[
LagTransform(in_column="target", lags=list(range(21, 57, 7)), out_column="lag"),
],
model=model,
horizon=HORIZON,
)
_, forecast_df, _ = pipeline.backtest(ts=ts, metrics=[MAE()], n_folds=3, forecast_params={"return_components": True})
plot_forecast_decomposition(forecast_ts=TSDataset(df=forecast_df, freq="D"), mode="joint", columns_num=2)
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.1s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.2s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.2s finished
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.1s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.1s finished
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.0s finished
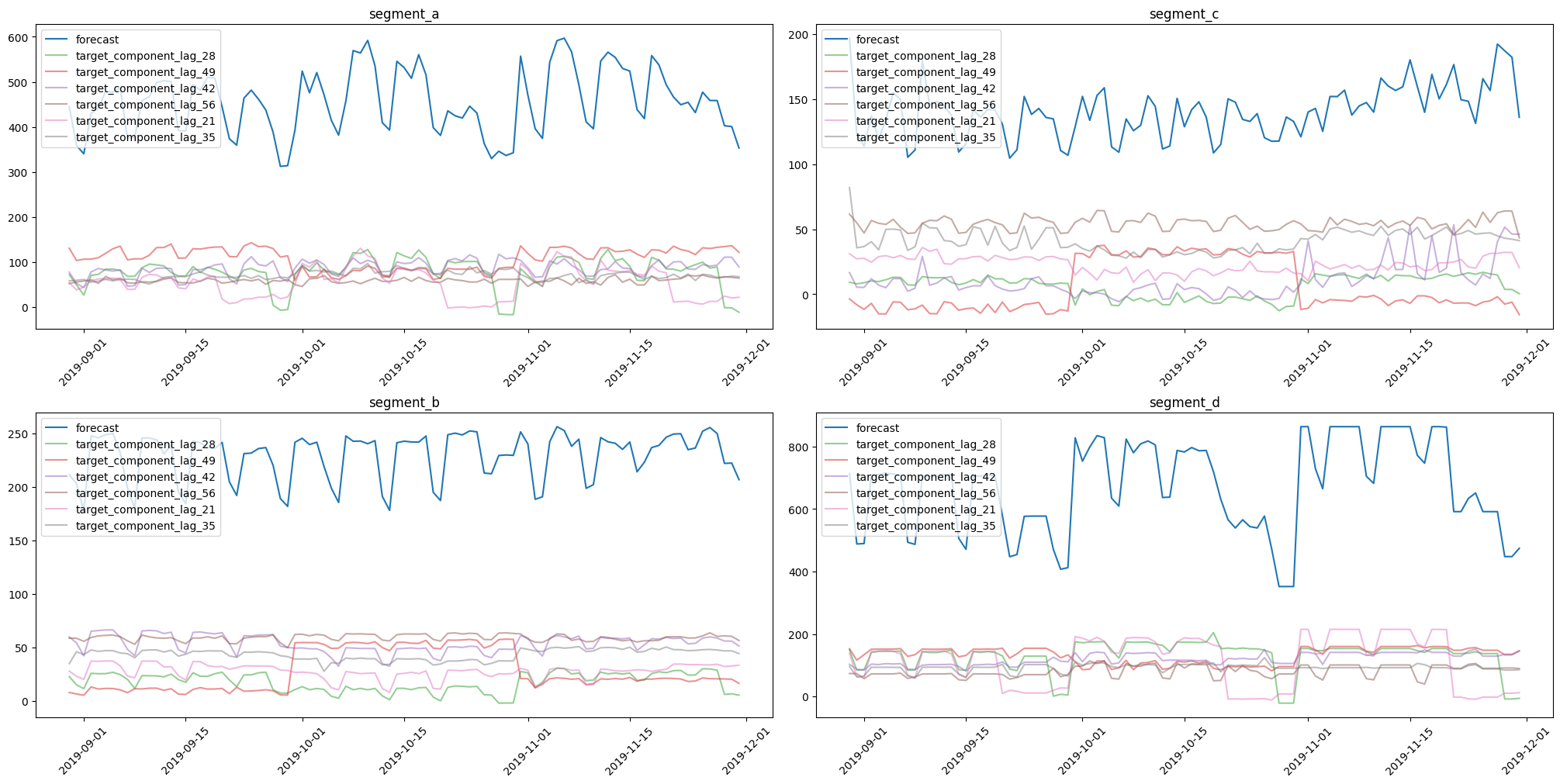
Creating a new model using sklearn interface¶
Now let’s create our model by leveraging already existing etna classes: * etna.models.SklearnPerSegmentModel: accepts sklearn-like model and creates etna-model that fits one model per each segment. * etna.models.SklearnMultiSegmentModel: accepts sklearn-like model and creates etna-model that fits one model on entire dataset & mdash; it is that we implemented in a section above.
[30]:
from etna.models import SklearnPerSegmentModel
from etna.models import SklearnMultiSegmentModel
First, let’s implement etna-model that fits separate model per each segment.
[31]:
class LGBMPerSegmentModel(SklearnPerSegmentModel):
def __init__(
self,
boosting_type="gbdt",
num_leaves=31,
max_depth=-1,
learning_rate=0.1,
n_estimators=100,
**kwargs,
):
self.boosting_type = boosting_type
self.num_leaves = num_leaves
self.max_depth = max_depth
self.learning_rate = learning_rate
self.n_estimators = n_estimators
self.kwargs = kwargs
model = LGBMRegressor(
boosting_type=self.boosting_type,
num_leaves=self.num_leaves,
max_depth=self.max_depth,
learning_rate=self.learning_rate,
n_estimators=self.n_estimators,
**self.kwargs,
)
super().__init__(regressor=model)
class LGBMMultiSegmentModel(SklearnMultiSegmentModel):
def __init__(
self,
boosting_type="gbdt",
num_leaves=31,
max_depth=-1,
learning_rate=0.1,
n_estimators=100,
**kwargs,
):
self.boosting_type = boosting_type
self.num_leaves = num_leaves
self.max_depth = max_depth
self.learning_rate = learning_rate
self.n_estimators = n_estimators
self.kwargs = kwargs
model = LGBMRegressor(
boosting_type=self.boosting_type,
num_leaves=self.num_leaves,
max_depth=self.max_depth,
learning_rate=self.learning_rate,
n_estimators=self.n_estimators,
**self.kwargs,
)
super().__init__(regressor=model)
Let’s try to recreate results of LGBMModel using LGBMMultiSegmentModel.
[32]:
model = LGBMMultiSegmentModel(random_state=42)
pipeline = Pipeline(model=model, transforms=transforms, horizon=HORIZON)
metrics_df_multi_segment, forecast_df, _ = pipeline.backtest(ts=ts, metrics=[MAE()], n_folds=3)
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.2s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.5s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.9s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.9s finished
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.1s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.3s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.5s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.5s finished
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.0s finished
Let’s look at the results.
[33]:
plot_backtest(forecast_df=forecast_df, ts=ts, history_len=50)
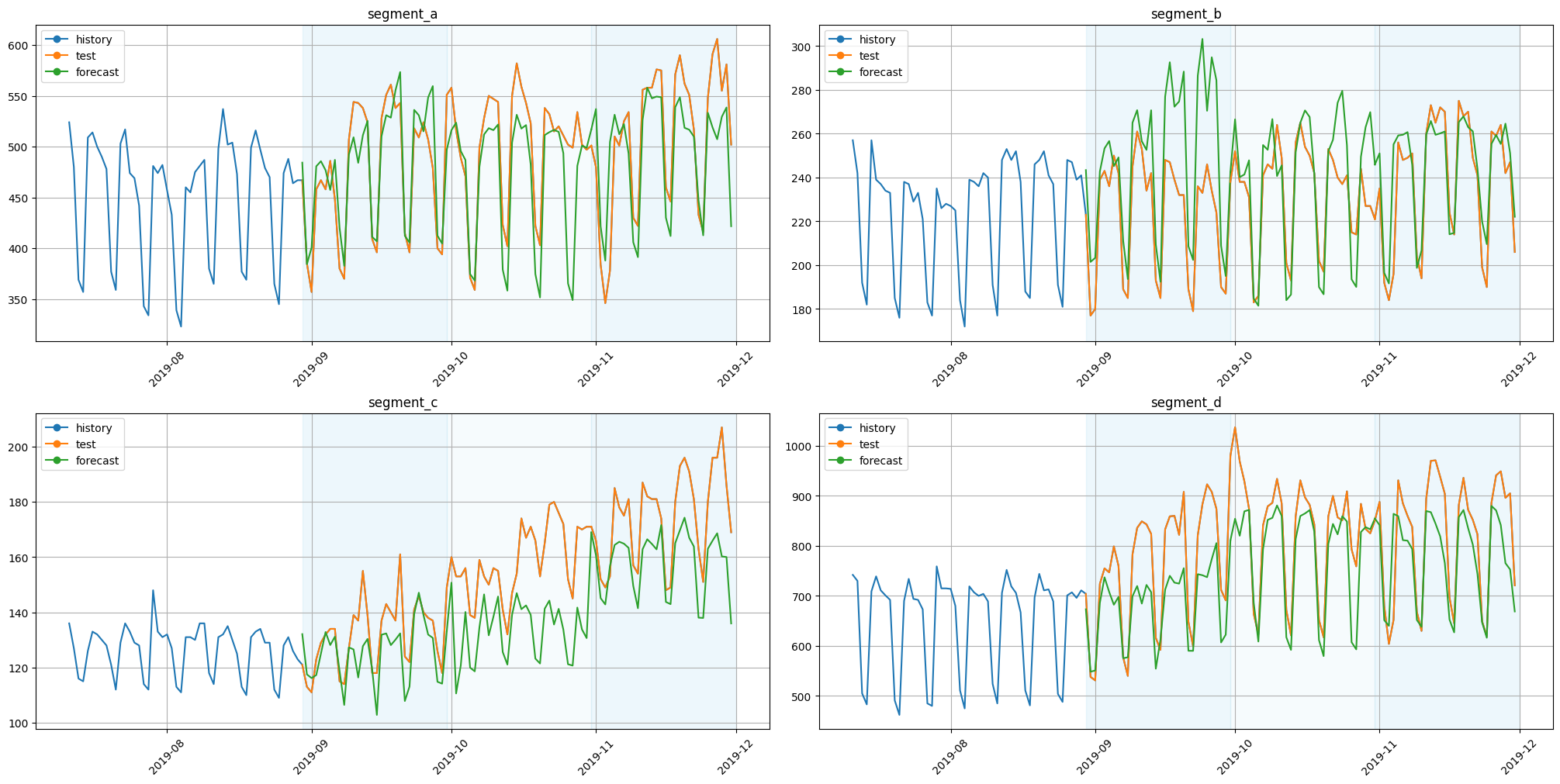
As we can see, the results are a little bit different. Let’s check this manually by looking at the values.
[34]:
metrics_df.head()
[34]:
| segment | MAE | fold_number | |
|---|---|---|---|
| 0 | segment_a | 22.633598 | 0 |
| 0 | segment_a | 35.034894 | 1 |
| 0 | segment_a | 35.003214 | 2 |
| 1 | segment_b | 22.285198 | 0 |
| 1 | segment_b | 16.546921 | 1 |
[35]:
metrics_df_multi_segment.head()
[35]:
| segment | MAE | fold_number | |
|---|---|---|---|
| 0 | segment_a | 23.121324 | 0 |
| 0 | segment_a | 34.062925 | 1 |
| 0 | segment_a | 34.721675 | 2 |
| 1 | segment_b | 25.587469 | 0 |
| 1 | segment_b | 13.960903 | 1 |
Why do we see this difference? In LGBMModel we have a special handling of categorical features, but in LGBMMultiSegmentModel we doesn’t have it, because etna.models.SklearnMultiSegmentModel doesn’t implement this logic with categorical features.
As you can see, etna.models.SklearnPerSegmentModel and etna.models.SklearnMultiSegmentModel have some limitations, but they should cover a lot of cases.
This raises a question: what if I want to implement per-segment logic manually with handling categorical features like in LGBMModel? A good reference for such a task will be the implementations of etna.models.CatBoostPerSegmentModel and etna.models.CatBoostMultiSegmentModel. There we use special mixins for per-segment/multi-segment logic.
If you want to add you model to the library don’t forget to write the necessary tests and documentation. Good luck!